
Only a tiny fraction of RNAs have experimentally determined structures.
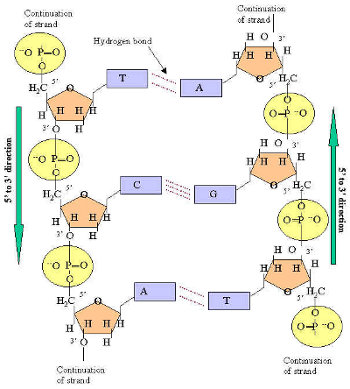
However, these methods have low throughput. RNA secondary structure can be determined from atomic coordinates obtained from X-ray crystallography, nuclear magnetic resonance (NMR), or cryogenic electron microscopy ( 11–13). As evidence of this, many homologous RNA species demonstrate conserved secondary structures, although the sequences themselves may diverge ( 10). For many RNA molecules, the secondary structure is essential for the correct function of the RNA, in many cases, more than the primary sequence itself. The base-paired structure is often referred to as the secondary structure of RNA ( 9). RNA folding is in large part determined by nucleotide base pairing, including canonical base pairing-A–U, C–G and non-Watson–Crick pairing G-U, and non-canonical base pairing ( 7, 8). The microRNAs are abundant in many mammalian cell types, targeting ∼60% of genes ( 5), and are often regarded as biomarkers for diverse diseases ( 6).Ĭellular RNA is typically single-stranded. The spliceosome, which performs intron splicing, is assembled from several snRNAs. Some RNAs possess catalytic functionality, playing a role similar to protein enzymes. Aside from its conventional role as an intermediate between DNA and protein, cellular RNA consists of many other functional classes, including ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), microRNA and other noncoding RNAs ( 1–4). The biology of RNA is diverse and complex. An online web server running UFold is available at. Its prediction is fast with an inference time of about 160 ms per sequence up to 1500 bp in length. UFold is also able to predict pseudoknots accurately. It significantly outperforms previous methods on within-family datasets, while achieving a similar performance as the traditional methods when trained and tested on distinct RNA families. We benchmark the performance of UFold on both within- and cross-family RNA datasets. UFold proposes a novel image-like representation of RNA sequences, which can be efficiently processed by Fully Convolutional Networks (FCNs).


Here, we propose a deep learning-based method, called UFold, for RNA secondary structure prediction, trained directly on annotated data and base-pairing rules. Traditional RNA secondary structure prediction algorithms are primarily based on thermodynamic models through free energy minimization, which imposes strong prior assumptions and is slow to run. Predicting RNA secondary structure from nucleotide sequences is a long-standing problem in genomics, but the prediction performance has reached a plateau over time. For many RNA molecules, the secondary structure is essential for the correct function of the RNA.


 0 kommentar(er)
0 kommentar(er)
